Theme1 Simulations of biomolecules in cellular environments
Simulations of biomolecules in cellular environments

Group Leader:
Yuji Sugita
(Theoretical Molecular Science Laboratory, RIKEN)
Multi Scale Modeling of Chromatin and Nucleosomes
Summary
By developing molecular-level simulation methods and carrying out research on potential applications, we are working to elucidate biological phenomena at the cellular level, and by doing to make possible new therapies and develop new drugs.
Most of the inside of a cell is water, but proteins actually take up 30%, and are constantly rubbing up against one another.
Many diseases begin when a protein is no longer able to fulfill its proper function. Because we understand this, a lot of modern research aims to get an accurate picture of the behavior of proteins in the cell. Analysis of the behavior was in practice impossible with the relatively slow computers of the past. We were limited to modeling the behavior of a single protein within a solution.We would like to look at a lot of proteins under conditions close to the cellular environment on an unprecedented scale with the excellent computational capacity of the K computer.
[lower Movie]
Double-stranded DNA in which genetic information is encoded is folded into compact protein-DNA complex structures, called “chromatin", in a nucleus of cell. When DNA is transcribed into RNA for gene expression, chromatin has to be in relaxed conformations. These conformational changes are regulated by chemical modifications of histones and so on. To study the complicated mechanism of life, three-dimensional structures of nucleosomes that compose the chromatin were constructed virtually and molecular dynamics (MD) simulations based on physical laws such as the equation of motion were conducted using the K computer. Such MD calculations enable us to simulate and observe dynamic behaviors of chromatin structures precisely.
Approach during the Past Five Years and its Achievements
Publications
1. Doki S, Kato HE, Solcan N, Iwaki M, Koyama M, Hattori M, Iwase N, Tsukazaki T, Sugita Y, Kandori H, Newstead S, Ishitani R, Nureki O:
“Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT“
Proc Natl Acad Sci U S A, 110 11343-8 (2013)
2. Jaewoon Jung, Takaharu Mori, Yuji Sugita:
“Efficient lookup table using a linear function of inverse distance squared“
J. Comput. Chem., 34, 2412-2420 (2013)
3. Michael Feig, Yuji Sugita:
“Reaching new levels of realism in modeling biological macromolecules in cellular environments“
J. Mol. Graph. Model., 45, 144-156 (2013)
4. Jaewoon Jung, Takaharu Mori, and Yuji Sugita:
“Midpoint cell method for hybrid (MPI+OpenMP) parallelization of molecular dynamics simulations“
J. Comput. Chem., 35, 1064-1072 (2014)
5. Kono H, Shirayama K, Arimura Y, Tachiwana H, Kurumizaka H:
“Two arginine residues suppress the flexibility of nucleosomal DNA in the canonical nucleosome core“
PLoS ONE.10:e0120635. (2015)
6. Naoyuki Miyashita, Suyong Re, Yuji Sugita:
“REIN: Replica-exchange INterface for simulating protein dynamics and function“
International Journal of Quantum Chemistry, 115, 325-332 (2015)
7. Michael Feig, Ryuhei Harada, Takaharu Mori, Isseki Yu, Koichi Takahashi, Yuji Sugita):
“Complete atomistic model of a bacterial cytoplasm for integrating physics, biochemistry, and systems biology“
Journal of Molecular Graphics and Modelling, 58, 1–9 (2015)
8. Yuki Shindo, Kazunari Iwamoto, Kazunari Mouri, Kayo Hibino, Masaru Tomita, Hidetaka Kosako, Yasushi Sako & Koichi Takahashi:
“Conversion of graded phosphorylation into switch-like nuclear translocation via autoregulatory mechanisms in ERK signalling”
Nature Communications 7, Article number:10485, (2015)
Group organization
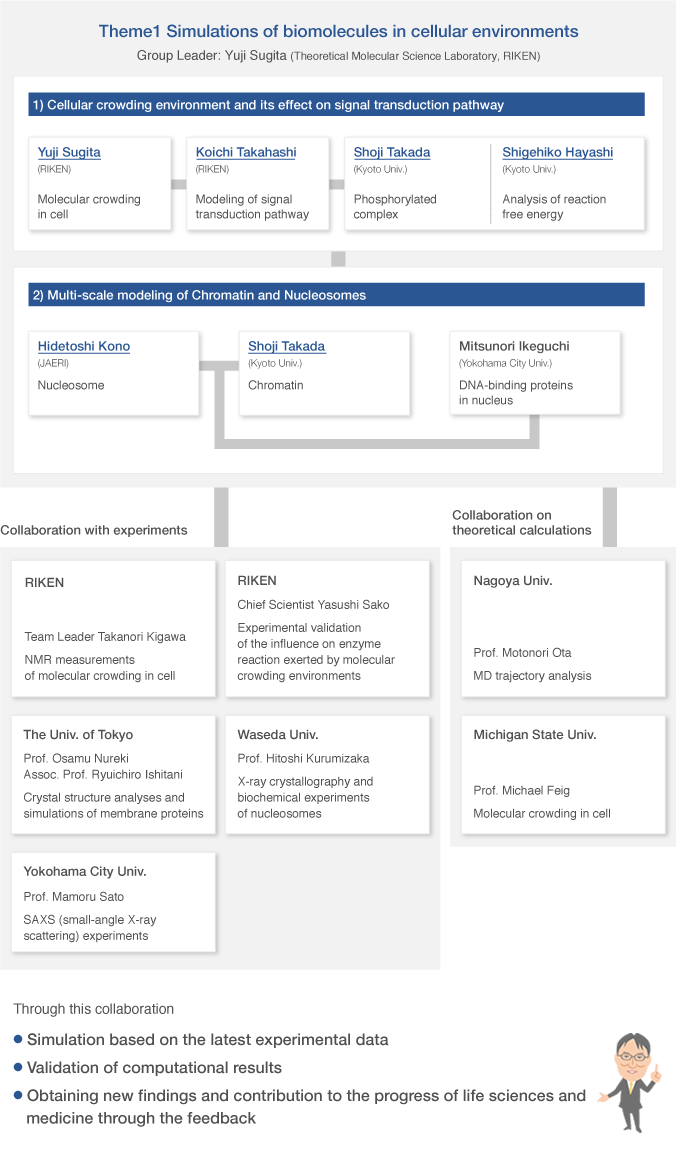
As of April 1, 2015