When you hear that “Water occupies approximately 70% of the whole space in a cell”, you may imagine an environment with sufficient space but actually, it is not. The space where 30% of the whole volume is occupied by molecules except those of water is extremely crowded environment, and a number of proteins or small molecules are packed in it like a crowded train. The question of how the proteins and small molecules move and interact each other in such a crowded environment is important for the fields of biology and drug design. However, it is difficult to measure the dynamics of molecules in cell with experimental ways, and therefore its details have not been clarified.
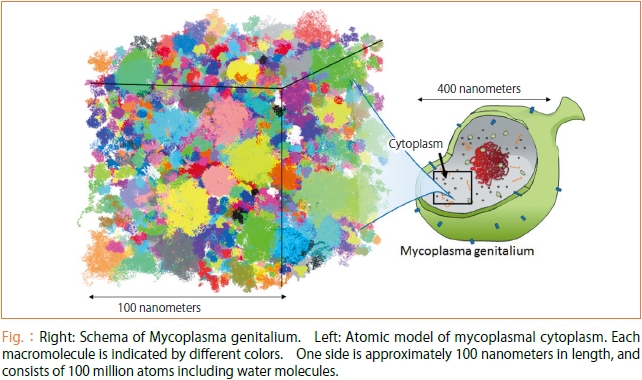
We aim to elucidate the dynamics of biomolecules in cell at the atomic level by large-scale molecular dynamics (MD) simulation using the K computer (simulation to reproduce the dynamics of molecules in detail by calculating forces that act on atoms, and repeatedly solving equations of motion for them) Specifically, we simulate cytoplasm of bacteria called Mycoplasma genitalium.
The constructed cytoplasm models consist of 10-100 million atoms. This ultra large scale model was developed by researchers of RIKEN (Takaharu Mori and Ryuhei Harada) and research collaborator in U.S. (Prof. Michael Feig, Michigan State University) [1]. Since all genes of mycoplasma have already been understood, the types and structure for most of the proteins can be estimated. Further, since omics data for other macromolecules and metabolites are available, we can also estimate types, structure, and concentrations of them in cell. In this way, we have successfully reproduced biomolecules (except cell membranes and DNAs), and their concentration in cell at the "atomic level", almost perfectly using various experimental data. We are simulating the "whole cytoplasm model" developed in this way with the MD simulation program "GENESIS" (developed by researchers of the Advanced Institute for Computational Science [2]) on the K computer. GENESIS is the program into which various new technologies are incorporated so as to enable super large-scale parallel MD simulation. Its efficiency does not decline even when hundreds of thousands of CPU's run simultaneously [3]. In other words, our study was made possible by combining the computation power of the K computer with the high efficiency of GENESIS.
Presently, the simulation itself has almost been completed, and now we are analyzing how the biomolecules move in cells within a time period of around 100 nanoseconds. We have confirmed that the speed at which proteins diffuse corresponds well with experimental values. In addition, we have obtained new insights for the structure of enzymes and dynamics of metabolites in cells, that can never be seen in a dilute solution environment in vitro. We intend its application to the field of drug design in the future, and are planning to perform longer simulations so as to investigate the interaction between proteins, and binding of metabolites and enzymes in cells.