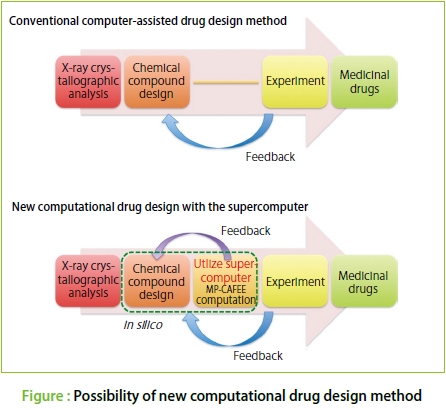
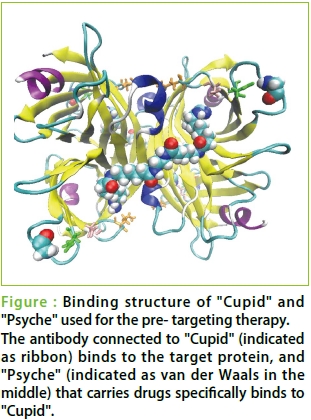
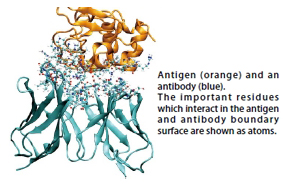
●First, I'd like to hear about how you two started working on the present research.
Yamashita : I have worked on the computational science since I was attracted to describing physical phenomena in mathematics in my high school days and worked on the computational science. I was interested in quantum mechanics and studied on proton. Having found a lot of phenomena greatly affected by the proton in the field, I was engaged in the biological field such as the molecular biology in the US and studied on proteins. That is my background. I was interested in drug design using protein simulation. I thought the simulation would be applied more widely than I expected, so I decided to challenge this theme. The research directly connected to medical service is related to priority issues in the society, so that is worth doing. I am really happy if I can contribute to the society. I joined the University of Tokyo in January, 2011. Since then I have studied on biosimulation and the molecular dynamics calculation that targets proteins related to drugs.
Shinoda : I was very attracted to the phenomenon called “phase transition” in physics in my college days. So, I worked in an experiment laboratory first, and was involved with studies mainly with X-ray diffraction experiments of alloy using synchrotron radiation. I had a chance to observe phenomena that could rarely be seen in the experiments and I was very excited with it. But, at the same time I hoped to dig into model researches and theory to find how and why such phenomena occur. I took a master course and a doctoral course of different graduate schools. I studied on phase transition of diluted ferromagnet in the master's course and on quantum liquid based on an integral equation theory in the doctoral course. I was involved with a research with the MD simulation for the first time when I was a post-doctoral researcher of National Institute of Advanced Industrial Science and Technology. After that, I had worked on various themes such as material development or quantum chemical calculation as a post-doctoral researcher of enterprises. Meanwhile, I had been in half-hearted attitude all the time. I could take time to hypothesize and carefully by myself. I could not dig into my research, which made me frustrated. I was employed by the University of Tokyo in December, 2010, a month before Dr. Yamashita was. With MD simulations of biomolecules using the supercomputer and examination of their structures and dynamics, I researched to develop drugs. Because the researches including examination of their structures and dynamics to support drug design have its clear purposes and direct relation to the output. Such researches really interested me and were worthwhile doing. The professors of the wet team working with me are eager to develop drugs with the results of computational science. The results of simulation research are directly applied for drug design, which gave me a great pressure on me; however, I will make an effort to do it.