-- When did you start the development of "GENESIS" ?
Sugita
We started it around 2010, before the emergence of the K computer. The computational method to simulate motions of molecules according to the laws of physics is called the "Molecular Dynamics (MD) method". A number of MD simulation software has been developed all over the world so far. If you try to apply conventional computational algorithms to a large-scale molecular assembly system with a large number of CPUs like the K computer, the computation speed is easily saturated due to the significant communication between CPUs. Therefore, we started to develop a new MD simulation program that is capable of fully utilizing the maximum performance of the K computer.
However, to be honest, it was obviously not easy to develop such software, and barriers to start the development were really high. First of all, we struggled to make MD software that can work on the K computer. However, unfortunately, we could not get the expected performance at first. Moreover, in order to create software that can be used for research, various functions should be introduced. I often felt it was a hopeless task during the development. I was afraid that Dr. Mori and Dr. Jung, who played a key role in the development, had to continue their work without clear prospects. We had no idea when we could reach the goal.
Mori
First, we made efforts to reproduce the same energy and force values as existing software. If we cannot obtain the same results, it means "GENESIS" has errors somewhere. After that, simulation methods such as temperature and pressure control algorithms were introduced. The algorithm that I introduced first was actually wrong. The temperature control was okay, but there were a lot of troubles in pressure control. Anyway, we solved the problems one by one while keeping studying. Because such basic algorithms are prerequisites for research, we planned to do basic work prior to developing new methods.
The good thing was that I like programing. The development was not so painful for me.

Jung
I had been involved in quantum mechanical/molecular mechanical (QM/MM) calculations before I joined this project. So I didn't know much about MD simulation. I had to learn about the theories of MD one by one, and wrote the source codes from scratch. It was really attractive for me to use the K computer, one of the most advanced computers in the world. This was my motivation to develop "GENESIS". I was sure that I would have a chance to experience new things I had never done. I expected to make new achievements, and thought that the new research topic was a good challenge for me.
Sugita
For the first 3 years or so, we could not write any research papers, because we have concentrated on the development.
-- When did you overcome the barriers,
and when did the development work with great progress?
Sugita
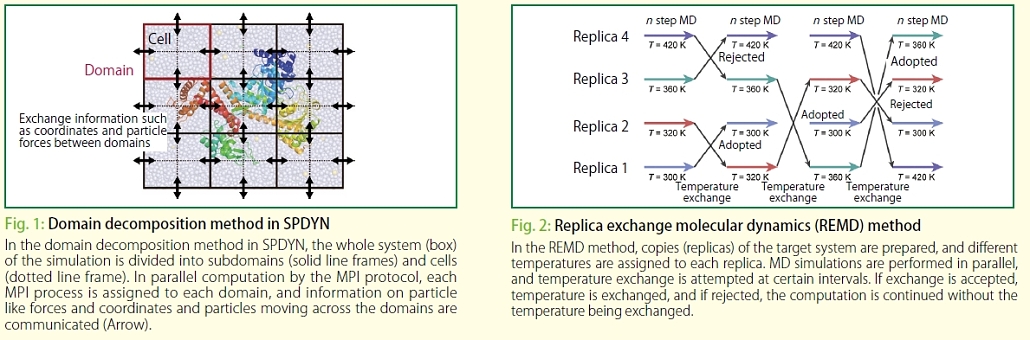
I think it was since the development of SPDYN got off the ground. "GENESIS" consists of two MD simulators named ATDYN and SPDYN, and analysis tools. In ATDYN, where the atomic decomposition method is introduced, interactions to be calculated are distributed over processors for parallelization. On the other hand, SPDYN uses an algorithm that divides the entire space into small subdomains and cells by the space decomposition method, and mainly communicates data between neighboring subdomains. In SPDYN, each subdomain is assigned to a processor, and interactions are computed in each subdomain. In other words, ATDYN divides interactions simply by the number of CPUs, while SPDYN divides data according to the space. Since, data communication is performed mainly between neighboring domains in SPDYN, communication costs are greatly reduced. Therefore, MD simulation could be performed faster for larger systems with SPDYN. We obtained efficient data partitioning by excluding unnecessary data. Because various techniques are introduced into SPDYN, the source codes are very complicated. To develop SPDYN, I guess that there had been a lot of problems, but Dr. Jung did a really great job.
-- Another important function of "GENESIS" is the Replica-Exchange Molecular Dynamics method (REMD), isn't it?
Sugita
In the REMD method, simulations of different conditions are mixed by preparing a number of copies of the target system. For example, simulations at different temperatures are performed in parallel, and temperatures are exchanged between replicas at a certain frequency. Exchanging temperatures enable us to search various structures of proteins in the simulations. Of course, the conventional MD may be able to do a similar structure sampling, but it requires much longer computational time. Presumably, it is difficult even with the K computer. In this respect, the REMD method enables us to obtain a lot of structures in the same computational time. We can analyze the mean value of physical quantities in those structures. More accurate calculations are possible. In the conventional MD, proteins often stay in states close to the initial structure at low temperature, while REMD helps us to search protein conformations in different states by mixing high temperature simulations. In addition, "GENESIS" is capable of changing various parameters such as pressure, surface tension and biasing forces as well as temperature. Dr. Mori worked on the development of the REMD method with great efforts.
-- "GENESIS" was released as open-source software from May 8, 2015.
Sugita
Because GENESIS has been updated frequently, we have to choose the best timing of release carefully. GENESIS has the top benchmark performance in the world, and we published a paper about it. At that time, we thought it was the best timing for the first release. Since our project is supported by tax, the products should be open, I think. GENESIS is GPLv2 license software. According to the license rule, third parties can modify the source code, and also put it for commercial use. If the third party distributes the software, the source code must be open. Even then, the software is free of charge. We hope that GENESIS will be used by not only academic researchers but also pharmaceutical companies. Although we have developed "GENESIS" for the K computer, it actually runs on most computers. We are going to add many other functions into GENESIS. Our future goal is to improve "GENESIS" up to the global standards of MD programs, in other words, the representative MD simulation software. In our laboratory, we expect that individual researchers add new functions to "GENESIS", and utilize them in their own research. Eventually, we will update "GENESIS" by incorporating those into the future release version.

Mori
I am now doing application research on membrane proteins using "GENESIS", and also developing "GENESIS" on the side. To do my application research, I have to introduce new algorithms into "GENESIS" that are not available currently. I think "GENESIS" should be developed gradually by doing this.
Jung
As for me, I’m developing an even faster and user-friendly "GENESIS" that can be used not only for the K computer but also for usual PC clusters. For example, the development of "GENESIS GPGPU version", where graphic processing units (GPU) are used for MD simulation, is going to be finished soon. It makes the MD simulation much faster. Furthermore, a project for the post-K computer has already started. "GENESIS" has been selected as one of the target applications of the post-K. A vendor and a software developer are jointly doing a co-design that allows evolution not only of the software but also of machine architecture at the same time so as to realize a faster "GENESIS".
--We expect that GENESIS will evolve due to an increase in the new users.

Sugita
Yes, I hope so., We intentionally developed two MD programs, ATDYN and SPDYN, in the GENESIS package. With SPDYN, we focused our efforts on increasing the performance. However, simplicity of the source codes was sacrificed. On the other hand, we made ATDYN very simple. Dr. Mori made great efforts for ATDYN. It's no exaggeration to say that ATDYN is the simplest MD program in the world. I think this feature of ATDYN is very important. If source codes of an MD program are not reasonably simple, young students and researchers do not try to read and understand them. We actually made ATDYN as simple as possible, because we want younger researchers to understand and modify the source codes by themselves. ATDYN can be used as an educational tool.
Mori
Yes I agree with you. I expect many people use ATDYN, because I developed ATDYN with the intention of creating the simplest MD program sacrificing the performance. There are lots of books about MD simulations, and we can learn theories of MD from them. But even so, it's not easy for MD beginners to make a program from scratch. In order to fully understand MD, various kinds of knowledge are necessary, and reading the source codes is often important. I recommend the beginners to see a simple program like ATDYN, and study how the MD simulation works.
Jung
I myself started the development by scrutinizing the source code of ATDYN written by Dr. Mori.
Sugita
Of course, both simple ATDYN and complicated SPDYN are easy to use, and they can be used in almost the same way with the same input data. Still, ATDYN is aimed at ease of use, whereas SPDYN is aimed at high performance.
--Just now, we said that "GENESIS" should become the world standard MD program. What are you going to develop "GENESIS" in the future to achieve that?
Sugita
As I already said, we designed the first "GENESIS" to realize high-speed calculation on the K computer, and we introduced basic MD functions into "GENESIS". In the future, as Dr. Mori just said, we hope to incorporate other useful functions that are not implemented in the current "GENESIS", and we are also planning to add original ideas of great worth, in other words, higher functionality. Another point is that we are trying to make "GENESIS" run faster on machines other than the K computer. The current "GENESIS" is well-suited to large-scale simulations using big computers like the K computer. However, it is not really suitable for small-scale simulations on usual computers. We also want to improve this situation in the future.
We believe "GENESIS" will be widely utilized for not only basic research but also industrial applications such as drug discovery.