For sustaining life, all living beings including human beings are undergoing diverse chemical reactions (i.e. metabolism) in their bodies. Metabolism is produced by transcription proteins which are synthesized (or expressed) based on the genetic information recorded in DNA. By expressing as many of those proteins as necessary when required, living beings strictly regulate their metabolism. Abnormalities in the mechanism for regulating the genetic expression constitute a factor for causing cancer or genetic disorders. To elucidate this mechanism, we are conducting research by using a molecular dynamics (MD) simulation which traces the structural change in proteins on a computer.
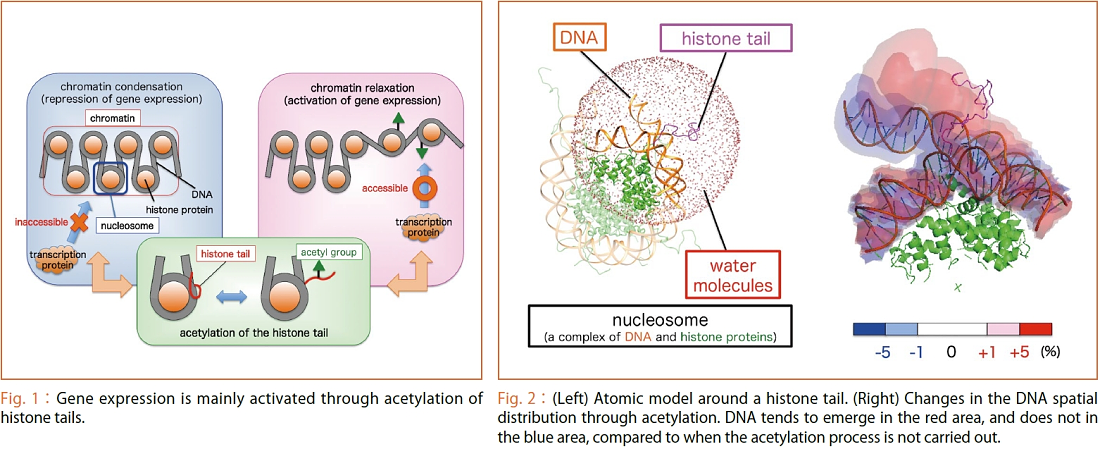
DNA is folded into compact structures in a cell nucleus. The structural unit formed by DNA wrapped around the histone proteins is called a nucleosome, and a fiber of packed nucleosomes called "chromatin". When the chromatin structure is condensed, transcription proteins cannot access DNA to decode the genetic information, and the gene expression is disabled. Meanwhile, if acetyl groups are added to histone tails (or terminals of histone proteins) (i.e. acetylation), the chromatin condensation is decreased, activating the gene expression (Fig. 1). To understand the mechanism for switching gene expression on and off, it is necessary to examine how the structures of histone tails change due to acetylation. However, as histone tails assume a number of structures called “intrinsically disordered states”, it is impossible to survey the molecular structures in detail using experimental methods such as crystal structure analysis or NMR spectroscopy.
Also, as histone tails carrying positive electrical charges strongly cling to DNA carrying negative electrical charges through electrical forces, conventional MD simulation has not identified the diverse structures of histone tails sufficiently.
As part of a project for the K computer, we succeeded in developing a new MD simulation method called adaptive lambda square dynamics (ALSD). This method allows us to examine diverse structures of histone tails by intentionally scaling their electrical charges in the simulation to eliminate the clinging to DNA. As a result, by using the computational power of the supercomputer, detailed pictures of histone tails, which had not been revealed through conventional experiments or simulated computations, emerged for the first time, such as the facts that the acetylation reduces the structural size of histone tails, leading DNA wrapped around histone proteins to relax easily and thereby allowing structural changes in chromatin to be promoted. (Fig. 2), and that acetyl groups are actively exposed over the surface of nucleosomes as a marker to induce transcription proteins. We will further clarify the gene expression mechanism and embark on research activities aimed at application such as the development of new drugs through the ALSD simulation method.
Visualizing the real molecular pictures, which are not identified through experimentation,
through the use of computing power
- Searching the structures of histone tails -
|
![]()
ZOOM IN Theme 3 Integrated simulations of the nervous system and musculo-skeletal system for reproducing Parkinson’s disease symptoms
ZOOM IN Theme 4 From supercomputing analysis of big cancer genome data to life science and medical care