The development of measuring techniques which enable non-invasive observation in the body such as MRI, X-ray CT and ultrasonic echo has contributed significantly to the advancement of medicine. Meanwhile, the development of computing science enabled dynamic phenomena to be analyzed at diverse dynamic layers from biological molecules and cells to tissues, organs and individuals, resulting in the reduction of the distances between biology, life sciences and physics.
One of the roles of bioengineering is to apply understanding of biological phenomena from such physical aspects to medicine. However, in the field of evidence-centered medicine, much importance is attached to measurements, and technical analyses based on mathematical models are not utilized sufficiently in clinical practice.
As the amount of data available because of progress in measuring devices increases and the target phenomena become more complicated, sophisticated diagnosis and treatment require deep understanding of the biological phenomena observed, and analysis of a huge quantity of measurement results. However, no medical engineering techniques to support such tasks have so far been developed.
The purpose of this Subtheme is to develop a computational simulator acceptable to the medical field where actual measurement data are important by assimilating and integrating large-scale physical simulations of the biological body through use of the post-K computer with biometric data supplied in various forms, and build fundamental technologies to support personalized medical care based on the medical and biological data of individual patients. For a large-scale simulator which can only be created with the post-K, we will develop a whole-brain circulation and metabolism simulator which connects cranial nerve activity with brain circulation. By providing the medical field with physical data obtained from the data assimilation based biological simulation, we will evaluate the biological functions in the human body correctly and quantitatively and build a database, and create new medical engineering technologies built on computational science in order to ensure sophisticated diagnosis, treatment, prevention and forecast methods which support a healthy, long-lived society.
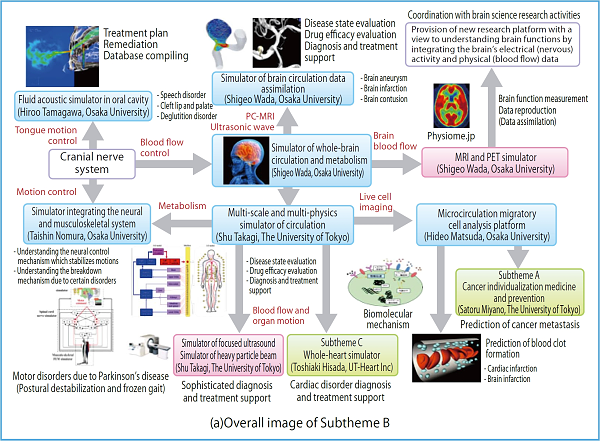
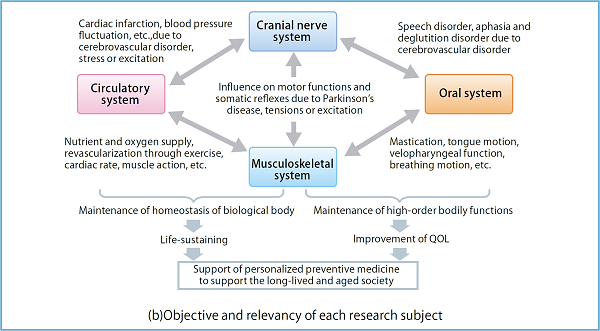
Fig. 1 : In Subtheme B, we will extend the simulation technologies developed by the K computer, and develop the biological body’s physical simulator based on each patient’s medical and biological data for the blood circulating system that plays the major role in life sustenance, the neural-musculoskeletal system responsible for body movements indispensable for the maintenance of QOL in the aging society, and the oral system responsible for speech.
Currently in the middle of transition from the K computer to the post-K computer, the “importance of planning research activities by forecasting progress in computers and the necessity for developing technologies for implementing this” are further increasing.
We are urged to think outside the box and steadily deepen our academic understanding.
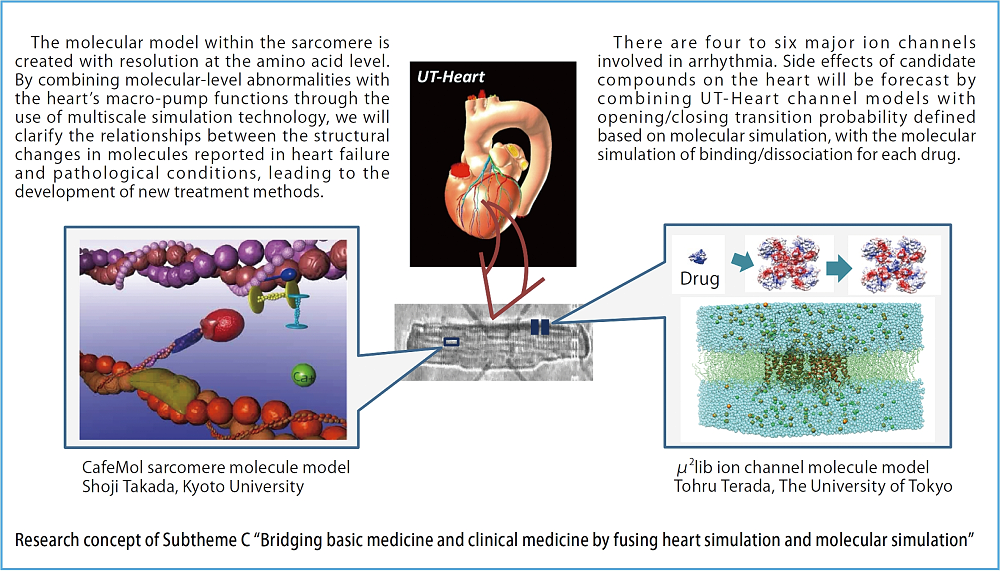
I would like to briefly present what we are going to achieve under the post-K project over the coming 10 years. The idea of our research activities is reflected in the subtheme, “Bridging basic medicine and clinical medicine by fusing heart simulation and molecular simulation.” In development activities by UT-Heart so far through the use of the K computer, multiscale simulation technology connecting micro contraction proteins which constitute the sarcomere with macro heartbeats was successfully developed. In this technology, a mathematical model, in which ATP is bound to the myosin head whose arm is expressed as a spring and undergoes hydrolysis, causing stochastic oscillations to/from actin filaments, is defined in accordance with statistical mechanics, and the resultant contraction force of the sarcomere is computed. For the post-K project, we will take this technology further to develop a sarcomere model consisting of coarse-grained molecular simulation models, and coordinate with the heart model. We consider that this will enable us to, for example, understand how the load observed and measured at the macro level is conveyed to the molecular level and activates the intracellular signal transducing system, and clarify the mechanism for causing pathological conditions. Moreover, by using the molecular model, we will
build the ion channel which controls the input/output into/from the cells of various ion currents which contain calcium ions that modulate the motion of contraction proteins, and simulate binding/dissociation to/from drugs, to evaluate the risk of arrhythmia caused by drugs to the heart. This will enable swift screening of candidate compounds which produce side effects on the heart and make the process of drug design more efficient. This study will be conducted in tandem with Priority issue 1.
Through good luck, we witnessed milestones, that is, the emergence of the K computer and subsequent evolution to the concept of the post-K. However, time passes so fast. Our achievements must be good enough to make constructive discussions during the next 10years. We would like to create a new milestone in the history of computing science by keeping our spirits high and concentration intense.
![]()
Priority issue (1) : Innovative drug discovery infrastructure through functional control of biomolecular systems
Subtheme A : Advancement of MD and associated algorithms by post-K computer
Subtheme B : Development of next-generation drug design computational techniques
Priority issue (2) : Integrated computational life science to support personalized ized and preventive medicine



















