Molecular dynamics simulation is commonly used as a method to find out the relationship between the molecular movement (dynamics) of macromolecules such as proteins and nucleic acids, and their functional structures. While conventional computations were targeted only at simulations under water or in lipid bilayer membranes, the objective of this Theme was to study molecular dynamics in a realistic cellular environment (biomolecules in cellular environments). At the initial stage of the study, we received criticism for ambiguity of specific computation targets. So we closely re-examined our research targets, and decided to focus on two research subjects: “modeling of signaling pathways considering cellular environments”, and “multi-scale modeling of chromatin and nucleosomes”. A research method termed “multi-scale simulation” played a fundamental role in this Theme. With the participation of specialists in quantum mechanics/molecular mechanics (QM/MM), all-atom molecular dynamics, coarse-grained molecular dynamics and single-particle simulation, biological phenomena from molecular to cellular scales were successfully analyzed by using molecular models with different resolutions. In particular, for the former, unprecedented large-scale simulations, such as all-atom molecular dynamics for bacteria cell cytoplasm, single-particle simulation for EGF signal pathways, and QM/MM computations taking the cellular environment into consideration, were conducted through the use of the K computer. For the latter, we succeeded in performing free-energy calculations on nucleosomes using a number of replicas, modeling of a tri-nucleosome through coordination between Small Angle X-ray Scattering (SAXS) and molecular dynamics ( MD / SAXS and CGMD / SAXS), large-scale chromatin modeling based on coarse-grained dynamics, and so forth. At this point, finally, the image of our initial objective, “biomolecules in cellular environments”, assumed a tangible form. I will continue to engage in my research activities by strengthening close cooperative relationships with other experimental researchers.

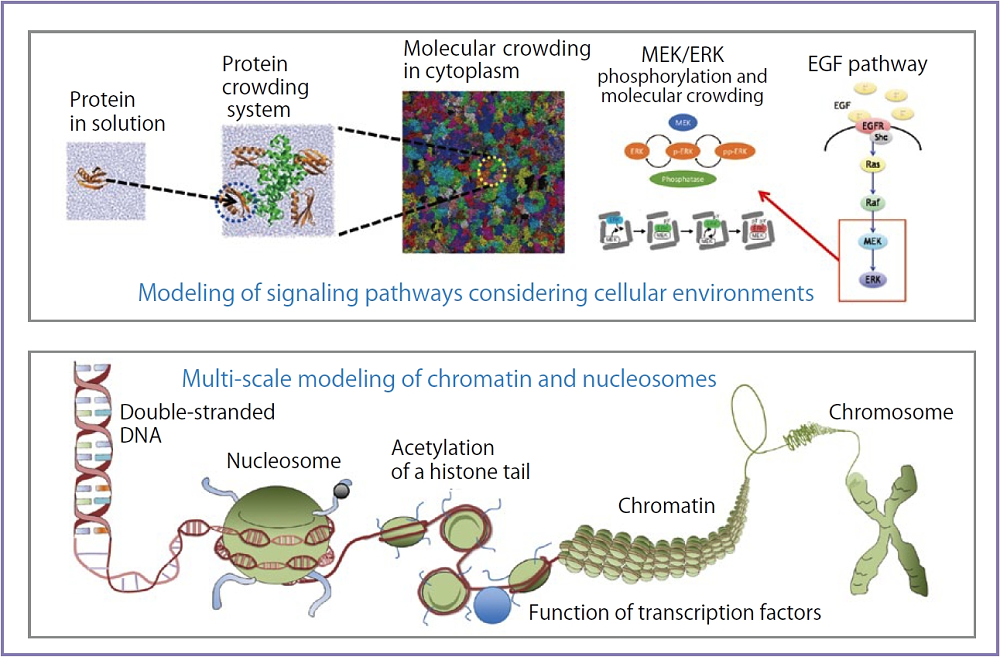
Fig. : “Modeling of signaling pathways considering cellular environments", and "multi-scale modeling of chromatin and nucleosomes”.
Open Up Special Talk : Approach to the four Themes during the past five years and its achievements
Theme 1 Simulations of biomolecules in cellular environments
Theme 2 Simulation applicable to drug design
Theme 3 Hierarchical integrated simulation for predictive medicine
Theme 4 Large-scale analysis of life data

















