
Computational Mechanobiology of Actin Cytoskeleton

Institute for Frontier Medical Science, Kyoto University
Yasuhiro Inoue (Cell Scale WG)
The actin cytoskeleton forms higher structures such as a network structure and bundle structure through interaction of various auxiliary proteins using actin filaments as the basic skeleton, in which actin monomers are coupled with one another, and constitutes an important mechanical and chemical base which supports cell shape and motion. For example, actin beneath the cell surface forms structures for motility such as filopodia and lamellipodia. They are essential for formation of neuronal growth cone and cell motion. In actomyosin filaments consisting of actin and myosin, the motor motion of myosin generates a force, which is crucial in cell deformation in morphogenesis, cytoplasmic division by a contractile ring, and turnover of cell adhesion. Those structures emerge from molecular systems centered on actin, and can change their structures and characteristics dynamically by going the round of mechanically and chemically metastable states. We are studying the relation of actin cytoskeleton dynamics to cell functions by using a computational mechanobiology approach. Our recent studies revealed that cell shape in cell motion is coupled with actin polymerization via thermal fluctuation [1], and the relation between the force generated in the actomyosin network and the myosin density is bilinear in response to network structure change [2]. These studies give important insights in the relation between cell-scale expression of function and molecularscale dynamics. Since actin mechanobiology can be simulated by using computers, it is not a totally unrealizable dream to recreate a whole cell on the computer. Of course, we have a lot of challenges to overcome. As for these challenges, we have to work with researchers inside and outside the Next-Generation Integrated Simulation of Living Matter Group. A Computational Science Algorithm Study Group is regularly held to create such an opportunity. There, researchers share knowledge of mathematical models and computational approaches beyond the scope of their fields, and pursue collaboration with researchers in related boundary areas.
【References】
[1] Inoue, Y and Adachi, T. Biomech. Model. Mechanobiol. 2011 10:495-503
[2] Inoue, Y. Tsuda, S., Nakagawa K., Hojo, M., Adachi T. J Theor Biol. 2011 21;281(1):65-73
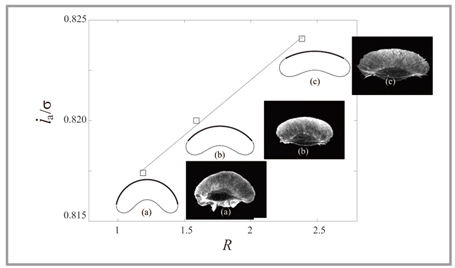 |
Fig 1 : Computational simulation of migrant cell: Cell shape obtained by simulation (solid line) coincides with actually-observed cell shape (fluorescence image). In addition, it was found that the actin polymerization speed at the tip of the cell (longitudinal axis) is in proportion to the curvature radius at the tip of the cell (horizontal axis). |
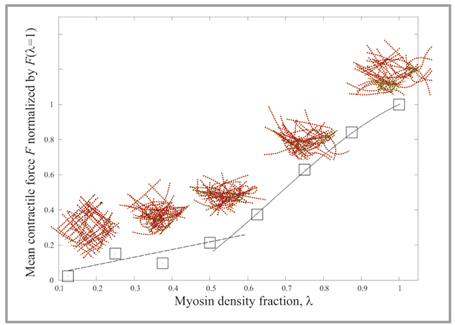 |
Fig 2 : Autonomous structural change of actomyosin net work : Due to motor motion of myosin in the actomyosin net work, the struc ture of ac tomyosin network changes autonomously. Simulation showed that the force generated in the network according to restructuring is bilinear with respect to myosin density. |
BioSupercomputing Newsletter Vol.5
- SPECIAL INTERVIEW
- The time has come for biosupercomputing to get results with the world's No. 1 supercomputer
"K computer", and take up the challenge of "prognostic biology".
Deputy Program Director of Computational Science Research Program, RIKEN Ryutaro Himeno - What should we do to promote industrial use of sophisticated computer resources and development applications?
Chief Coordinator of Foundation for Computational Science Masahiro Fukuda
Chief Researcher of Urban Innovation Institute and Executive Board Member and Bureau Chief of BioGrid Center Kansai Ryuichi Shimizu
- Report on Research
- Analysis of molecular mechanism of enzymatic reactions by QM/MM Free Energy Method
Graduate School of Science, Kyoto University Shigehiko Hayashi (Molecular Scale WG) - Computational Mechanobiology of Actin Cytoskeleton
Institute for Frontier Medical Science, Kyoto University Yasuhiro Inoue (Cell Scale WG) - Development of Blood flow Analysis Method for Simulation of Thrombus Formation
Department of Mechanical Engineering, The University of Tokyo Satoshi Ii (Organ and Body Scale WG) - Development of Data Assimilation Technology for Simulation of Living Things
The Institute of Statistical Mathematics Tomoyuki Higuchi (Data Analysis Fusion WG)
- SPECIAL INTERVIEW
- Pioneering the Future of Computational Life Science toward Understanding and Prediction of Complex Life Phenomena
Program Director of RIKEN HPCI Program for Computational Life Sciences Toshio Yanagida
Deputy- Program Director of RIKEN HPCI Program for Computational Life Sciences Akinori Kidera
Deputy- Program Director of RIKEN HPCI Program for Computational Life Sciences Yukihiro Eguchi
- Report on Research
- Simulation Applicable to Drug Design
Research Center for Advanced Science and Technology, The University of Tokyo Hideaki Fujitani (Field 1- Program 2) - An Ultra-fast Analysis System for Next-Generation DNA Sequencer Data
Graduate School of Information Science and Engineering, Tokyo Institute of Technology
Yutaka Akiyama, Takashi Ishida, Masanori Kakuta, Shuji Suzuki (Field 1- Program 4)
- Report
- BioSupercomputing Summer School 2011
Computational Science Research Program, RIKEN Yasuhiro Ishimine (Organ and Body Scale WG)
Research and Development Center for Data Assimilation, Institute of Statistical Mathematics Masaya Saito (Data Analysis Fusion WG)
Niigata University of International and Information Studies Eisuke Chikayama (Cell Scale WG)
School of Medicine, Tokai University Yohei Nanazawa (Cell Scale/Organ and Body Scale WG)
Computational Science Research Program, RIKEN Takashi Handa (Brain and Neural Systems WG)
Computational Science Research Program, RIKEN Gen Masumoto (High-performance Computing Team)
Computational Science Research Program, RIKEN Kei Moritsugu (Molecular Scale WG) - “Next-Generation Integrated Simulation of Living Matter (ISLiM)”, a web page dedicated to new applications, has opened.
Computational Science Research Program Integrated Simulation of Living Matter Group
- Event information
