Multi-scale simulation of platelet aggregation in initial stage of thrombogenesis

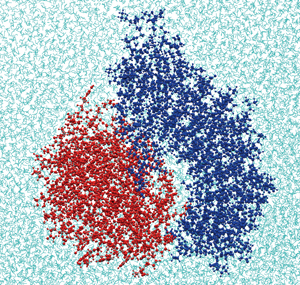
Fig. 1 :Binding site structure of N-terminal of platelet membrane protein GPIb alpha (blue) and A1 domain of VWF (red)
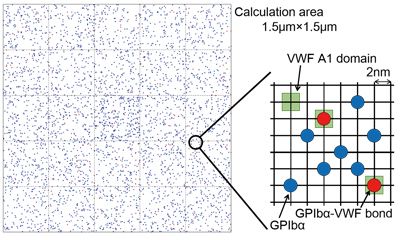
Fig. 2 :Platelet surface grid model. Red dots represent bound GPIb alpha, and blue dots represent free GPIb alpha which moves over the platelet surface.
When blood vessels are damaged by injury, etc., platelets clump together at the site of injury to stop bleeding. When the inner wall of a blood vessel is damaged due to arterial sclerosis, platelets also agglutinate and form a blood clot inside the blood vessel. Such blood clots cause thrombosis including heart infarction and cerebral infarction, which are major cardiocirculatory diseases and rank fourth as a cause of death in Japanese people. Platelets agglutinate in a blood vessel through interaction between glycoprotein Ib alpha (GPIb alpha) found on platelets and the von Willebrand factor (VWF) adhering to the wall of an injured blood vessel. A few dozen GPIb alpha-VWF couples are formed on the joining surface of the platelet to stick platelets together. On a larger scale, the dynamic action of erythrocytes in blood plays a crucial role in adhesion of platelets.
Fig. 1 shows the binding site structure of the GPIb alpha-VWF couple obtained by the molecular dynamics method. The molecular dynamics method can determine the momentary position of every atom by solving Newton’s equation of motion for all atoms constituting the protein molecules and water. By this method, we can calculate the force working between the proteins and the potential energy of the system we calculate. It enables us to model reaction rates of bond formation and dissociation of a protein couple. Fig. 2 shows a grid model of a platelet surface. In this model, the behavior of a membrane protein on the platelet surface is analyzed by a statistical approach called the Monte Carlo method to find the momentary bond number and adhesion force of the protein. In the calculation area, 1.5μm on a side, which is equivalent to about one fifth part of the platelet surface area, there are 4,000 GPIb alpha and 3,600 VWF binding sites. Each dot in Fig. 2 represents one GPIb alpha molecule. We presumed that GPIb alpha is scattered on the platelet surface whereas VWF is anchored to the blood vessel wall, and that GPIb alpha molecules bound to VWF are neither scattered nor move over the membrane. We also presumed that GPIb alpha near VWF forms a bond with VWF at a certain combination rate, and the bond is dissociated at a certain bond dissociation rate. As those rates, we can use a model modeled by the molecular dynamics method. By introducing the number of protein bindings and their adhesion force into an Eulerian approach for fluid-hyperelastic body interaction by the finite difference method, it becomes possible to reproduce a phenomenon in which platelets adsorb onto a blood vessel wall in the presence of erythrocytes [1].
By performing a multi-scale simulation from the protein scale to the bloodstream scale in this way, we are studying further to elucidate the clotting mechanism, and in the future we will construct a useful model for the discovery of an antiplatelet drug, and predictive and personalized medicine.
Reference:
[1] BioSupercomputing Newsletter, Vol. 7, p. 7.


















